In this month’s blog I caught up with NuNano co-founder and director Professor Mervyn Miles to discover not only where his fascination with microscopy and high-speed imaging came from, but also where he thinks the next exciting areas of research are to be found...
How did you get involved in microscopy?
It’s funny because I never set out to work with microscopes. My PhD actually focused on understanding polymer samples and for the first part of my career, understanding and predicting the structure of polymer samples remained my main point of study.
During my time at Birmingham University, I was granted access to a million electron volt (MeV) electron microscope.
Now in the normal course of things polymer samples tended to get destroyed after a couple of seconds in electron microscopes operating in the 100 keV range. Not with this one though!
Using the mega-volt microscope, I could see all sorts of pretty diffraction patterns. My sample looked almost as though it was on fire in dark-field imaging. I couldn’t believe what I was seeing, that it was possible to see this much detail without killing the sample.
From that point on I was hooked on microscopy and imaging - though at that stage only because it was a fascinating tool to facilitate my polymer research.
So a key part of your polymer research was around accessing the best possible imaging equipment?
Yes, you could say that. I went to Germany to do my post-doc in synthetic polymer physics studying under Herbert Gleiter[1]. He had a whole new way of looking at polymers, again using electron microscopes[2].
He was an amazing guy to work with, really inspirational. He had a constant stream of ideas and treated everyone as an equal despite the fact he was working on some complex stuff and cutting edge ideas. In many ways I internally absorbed his way and pattern of working.
During this time I obtained some pretty interesting images, showing the nanoscale structures of polymer fibres produced under elongational flow stretched single molecules[3]. I showed these some years later at a conference which was attended by Andrew Keller, a polymer physicist working at the University of Bristol. Keller was pretty impressed with what we'd achieved, which sorts of shows that it was ground-breaking stuff we were working on.
After I completed my post-doc in Germany I went to Case Western Reserve University, Cleveland, Ohio, which at the time had the best polymer department in the U.S. That was a totally different experience to working with Professor Gleiter. The guy I worked with in the Department of Macromolecular Science had great drive, but his focus and strength was in co-founding and building this department from the ground up, rather than the science. He pretty much delegated the running of his quite substantial research group to me. So I learned a lot quickly - many different projects needed data interpreting and new ideas and new directions.
After just one year in the U.S., I started my first period in Bristol. Prof Andrew Keller, who founded the field of polymer physics, had told me that if I ever wanted a job to contact him. It was time to cash in this offer! In that move, I did two very different jobs: transmission electron microscopy of polymers and elongational flow of polymer solutions to understand the nature of individual molecules in solution and what happen when they were stretched out by the flow field. This all went very well, but after three years it was time to find a ‘proper job’ rather than another post-doc position.
My next move, still focusing on polymer research, was to Norwich, to work – rather bizarrely – for the Institute of Food Research. They had loads of money to do basic science which in turn meant we weren’t constrained in the science we could do. Though ideally the sample should be edible!
Our head of division was a fascinating enigmatic guy called Henry Chan. He moved so quietly around the place he would suddenly appear at your shoulder whilst you were working and say, ‘you’re in trouble Miles, …. Big trouble’. I never found out if I really was or not. But brilliant. He was the person who first introduced me to and encouraged me to work with the new science of scanning tunnelling microscopy (STM).
Nobody really knew what it was about of course. Even at the European Bioscience Physical Congress, held in Bristol in 1984 – a huge conference with many parallel sessions including one on x-ray microscopy. I found myself making small talk with a chap needing help to find the building for the next session, I asked him was his area of research was. He told me about this esoteric technique where a sharp piece of wire is brought within an Angstrom or so of the sample surface and the quantum mechanical tunnelling current is measured as it is raster scanned over the sample. I thought this would never work because of the mechanical stability and control precision that would be needed. This was of course scanning tunnelling microscopy for which he, Heini Rohrer, and his colleague Gerd Binnig would be awarded the Nobel Prize in Physics two years later, and with the help of Prof. Sir Mark Welland in Cambridge, I began the change in direction in my career to this technique.
Through Dr. Chan’s and Dr. Morris’s encouragement I took one of the protein molecules I had been studying with small-angle x-ray scattering (SAXS) and deposited on an x-ray mirror’s surface, amorphous carbon, and looked at it via the STM with Mark Welland. It was pretty amazing. Through the images we obtained, we discovered the 3D structure the STM was showing us of individual molecules was as I had predicted from SAXS - we got one of the first pictures of a single protein molecule[4].
When did you start getting involved in the development of instrumentation?
By this point I was increasingly interested in the latest and best developments in microscopy. My focus was on improving the substrate for immobilising biomolecules rather than the tool itself though. I was always looking for the best substrate materials.
In 1989 I moved back to the University of Bristol, again working with Prof. Andrew Keller. I was supposed to be working on x-ray diffraction and scattering, but clearly STM was going to be huge, so I applied for an STM grant immediately I arrived in Bristol and we were awarded it in April 1990. This meant we finally had our own STM to play with!
Around this time one of my undergraduate project students built a scanning near-field optical microscope (SNOM) which achieved the best resolution of anywhere in the world and this is really where developing instruments came in for me, from around 1990. This is the photon analogue of STM.
Shortly after that, I put in for a grant and got an Atomic Force Microscope (AFM). I recruited a post-doc, Terry McMaster, from Norwich, and we began work on AFM of biomolecules.
So when did you start your first company?
When I was awarded a personal chair, that is, promoted to professor at Bristol, I decided to do something different. I began work on developing holographic tweezers and also started a company: Infinitesima.
We produced and sold a product called Activ Q which helps to control the quality of factor of the cantilever and is very important in liquid for improving image quality in liquid. We sold quite a few which was encouraging. One of the key issues for a while had been around improving imaging quality and stability in liquids.
We started trying out ideas to increase the speed of imaging, initially in SNOM, where we managed to increase the frame rate 100,000 times. We then wanted to try this for AFM, which would have far more applications.
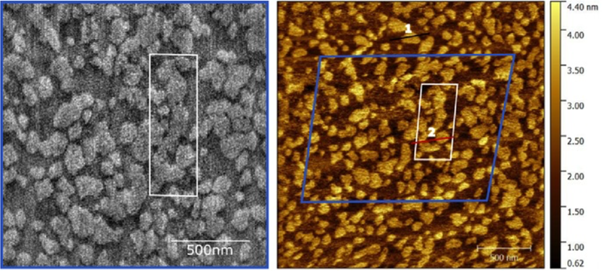
Amazingly it worked – and even more amazingly the speed was critical to it being able to produce great images in liquid (see figure below for a recent example of our high-speed AFM imaging). If we had turned the speed up slowly we probably wouldn’t have continued along this route because it turns out that at a slower speed the sample continues to be destroyed! Another one of those, ‘let’s give it a go and see what happens’ instincts that turned out well!
So where did the idea to set up NuNano come from?
It was about ten years after I’d started Infinitesima. Frustrated by the varying quality of existing AFM probes, we thought maybe there was an opportunity to make improvements. It was the brain child of my colleague and co-founder Heinrich Hoerber. The mix of what seemed to me to be a great idea with the enthusiasm and drive of former PhD student and post-doc James Vicary made setting up the company a no-brainer.
James has done a brilliant job of implementing the idea and turning it into a successful product. And, importantly developing out from that original idea to produce the kind of game-changing probes that no-one else is working on yet, such as the ultra-soft high-speed vertically-oriented probes (VOPs).
What excites you about the future of nanotechnology and what’s the next area of research you’re most interested in?
I think the use of high speed VOP force microscopy in true non-contact mode, with zero normal force, on living cells will make a major impact. The ability to see signalling, transport and whole changes in structure at the cell membrane will give exciting new information.
Using the high speed vertical probes for example, as I have done lately with work I’ve been doing around Alzheimer’s, just makes you realise how much more there still is to be explored in the world of force microscopy.
Imaging membranes at high speed means you don’t end up making holes in the lipid (and thus destroying the sample). We’ve produced the most amazing images of membrane samples where you can actually see the Amyloid proteins destroying the membranes.
It's exciting and important work and we're looking at ways to go beyond just imaging.
High-speed (left) and normal speed (right) AFM images of the same area of a model multi- lipid component neuronal membrane. The contrast corresponds to the slightly different height of each lipid. Such membranes are very soft, almost liquid-crystal-like, yet the disruption by the high-speed tip is surprisingly almost non-existent. Image courtesy of Morgan Robinson & Zoya Leonenko (University of Waterloo, Canada) and Loren Picco, Ravi Sharma & Mervyn Miles (University of Bristol, UK).
[1] H Gleiter, “Nanoglasses: A new kind of Noncrystalline Material and Way to an Age of New Technologies?” Small 12 (2016) 2225–2233
[2] J Petermann and H Gleiter, “Direct Observation of Amorphous And Crystalline Regions in Polymers by Defocus Imaging”, Philosophical Magazine 31 (1975) 929-934
[3] MJ Miles, J Petermann & H Gleiter, “Deformation Mechanism of ‘Hard’ Elastic Fibres’, Colloid & Polymer Science, 62 (1977) 6-8.
[4] ME Welland et al., "The Structure of the Globular Protein Vicilin revealed by Scanning Tunnelling Microscopy", International Journal of Biological Macromolecules, 11 (1989) 29-32.